Structure of MethSMRT
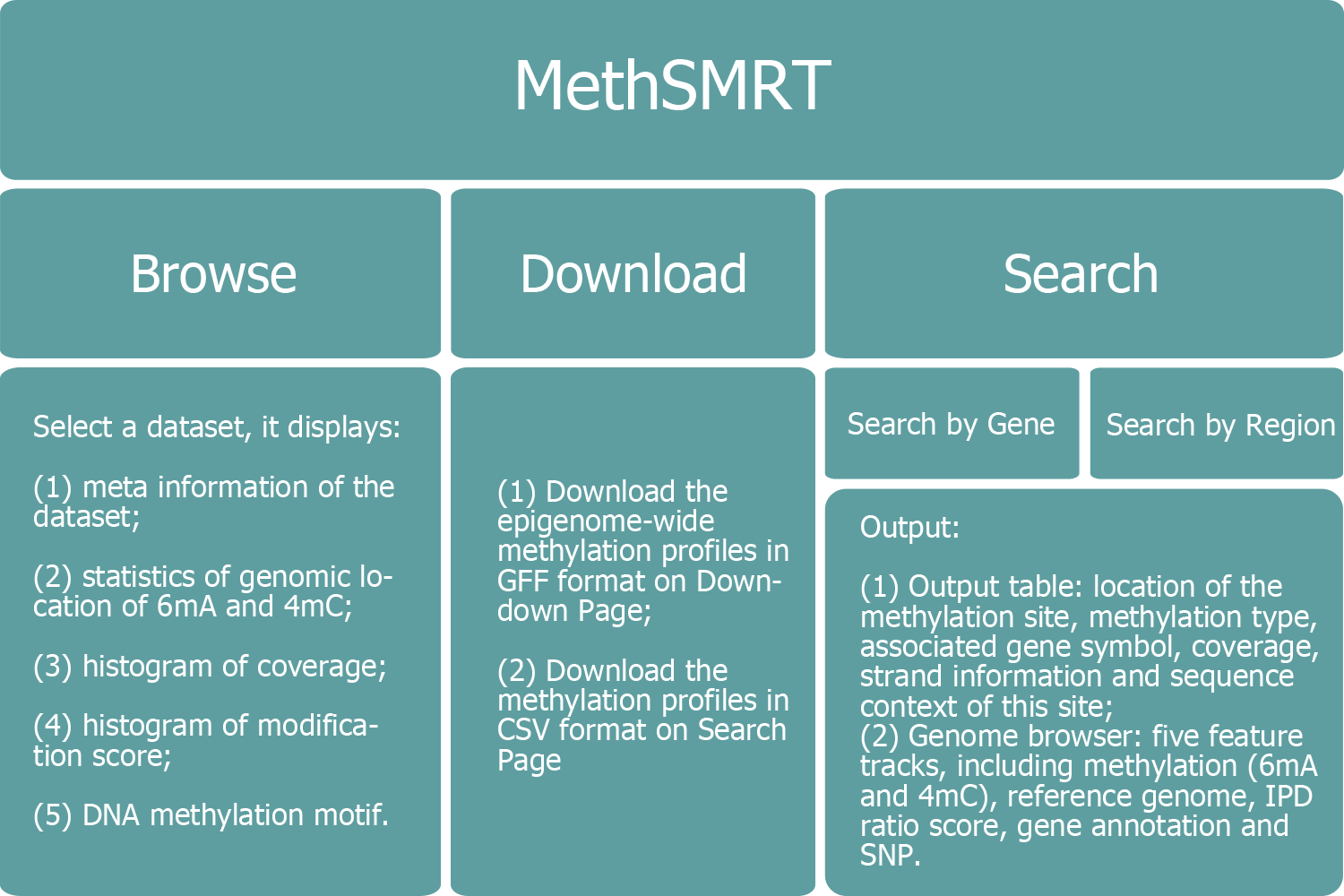
Home
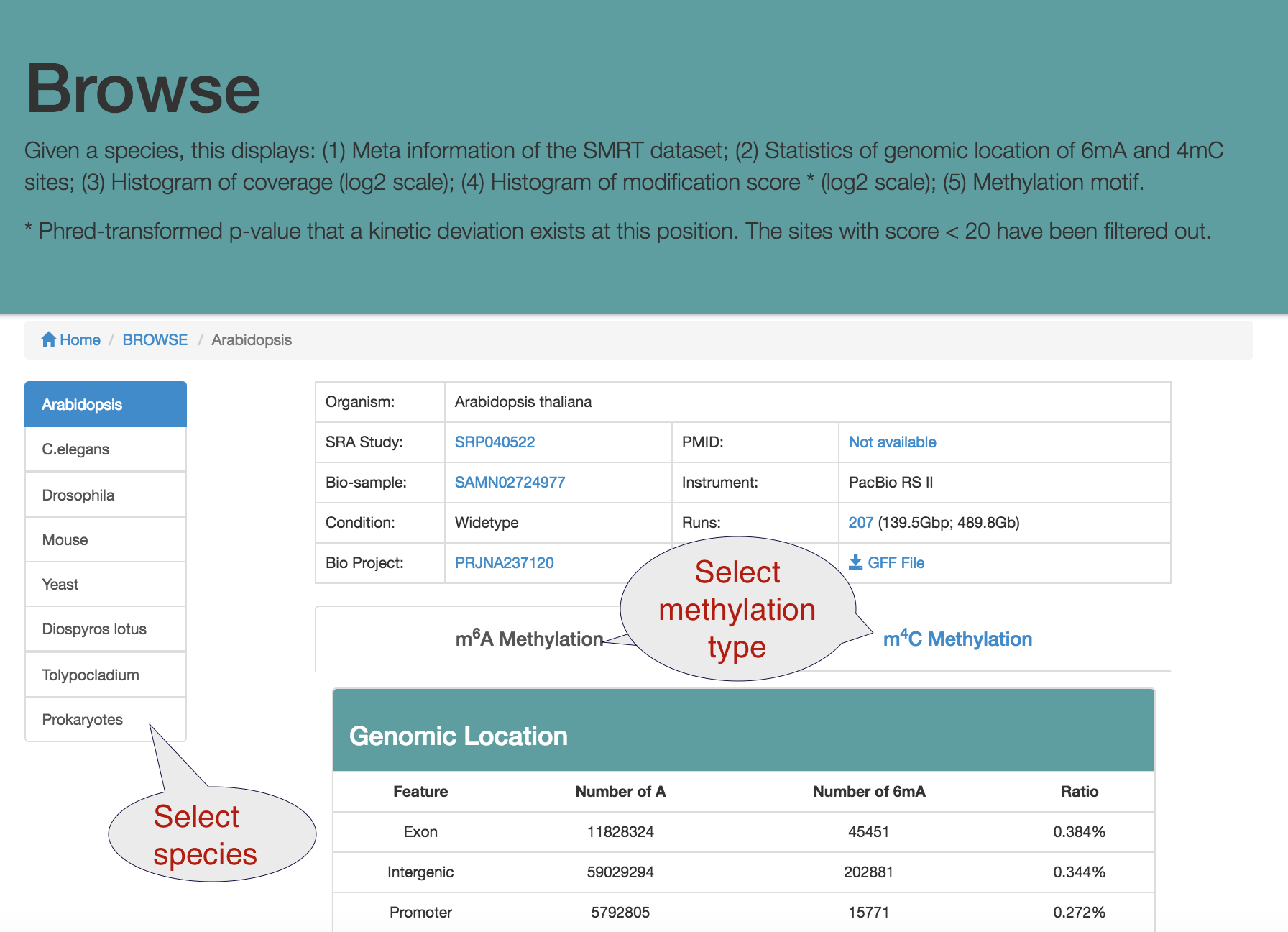
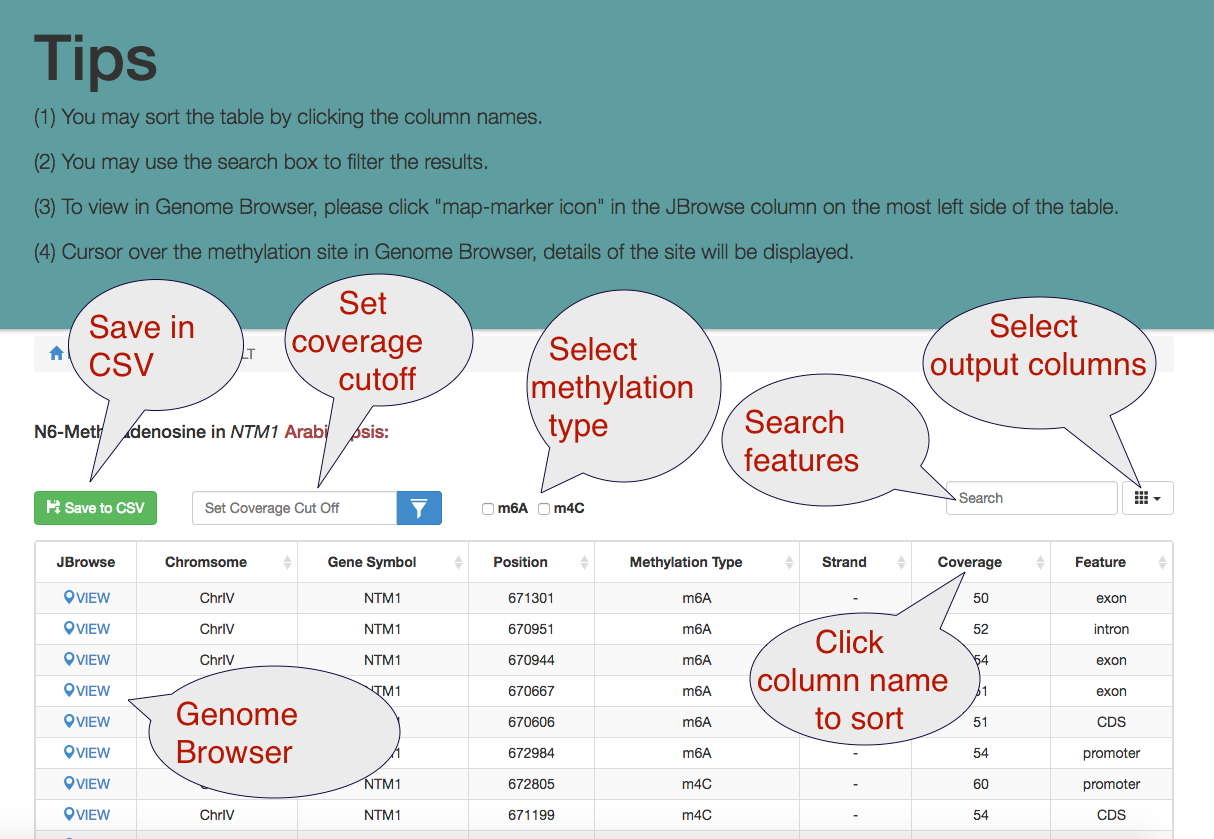
Browse
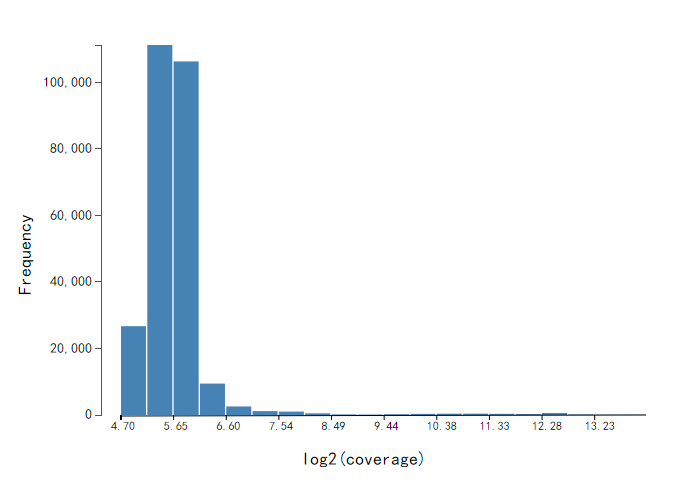
Histogram of coverage of methylation sites
X-axis indicates the coverage (number of reads) of methylation sites, in log2 scale. Y-axis indicates the number of methylation sites. The higher coverage, larger confidence with the identified methylation site. Followed the recommendations by the PacBio, the 6mA or 4mC sites with less than 25-fold coverage per strand were removed from further analysis.
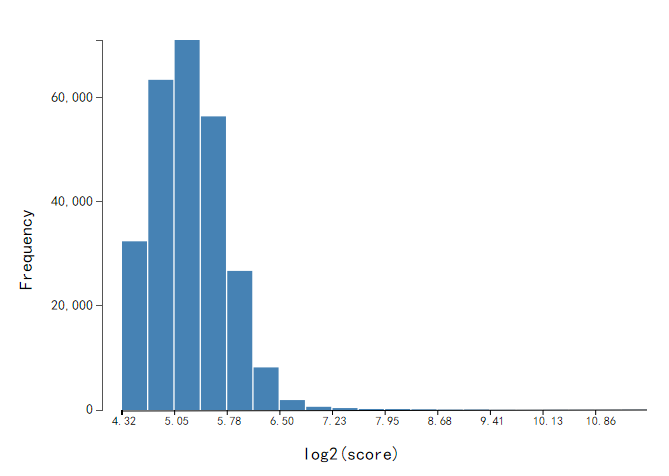
Histogram of modification score of methylation sites
The modification score is defined as phred-transformed p-value that a kinetic deviation exists at the position according to the PacBio manual. The higher score, more reliable the identified methylation site. X-axis indicates the modification score of methylation sites, in log2 scale. Y-axis indicates the number of methylation sites. To gain a reliable modification site, the sites with modification score < 20 have been filtered out from further analysis.
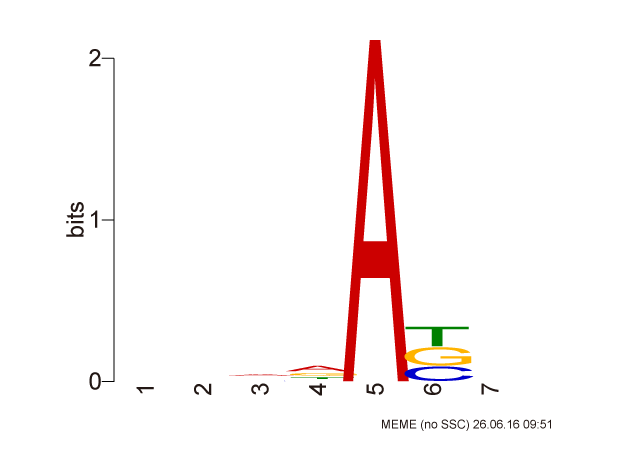
DNA motif of methylation sites
To infer DNA methylation motif, we first extracted sequences between 4-nt upstream to 4-nt downstream of the modification site. Duplicated sequences were excluded and MEME (version: 4.11.1) was used to predict enriched motifs.
Search
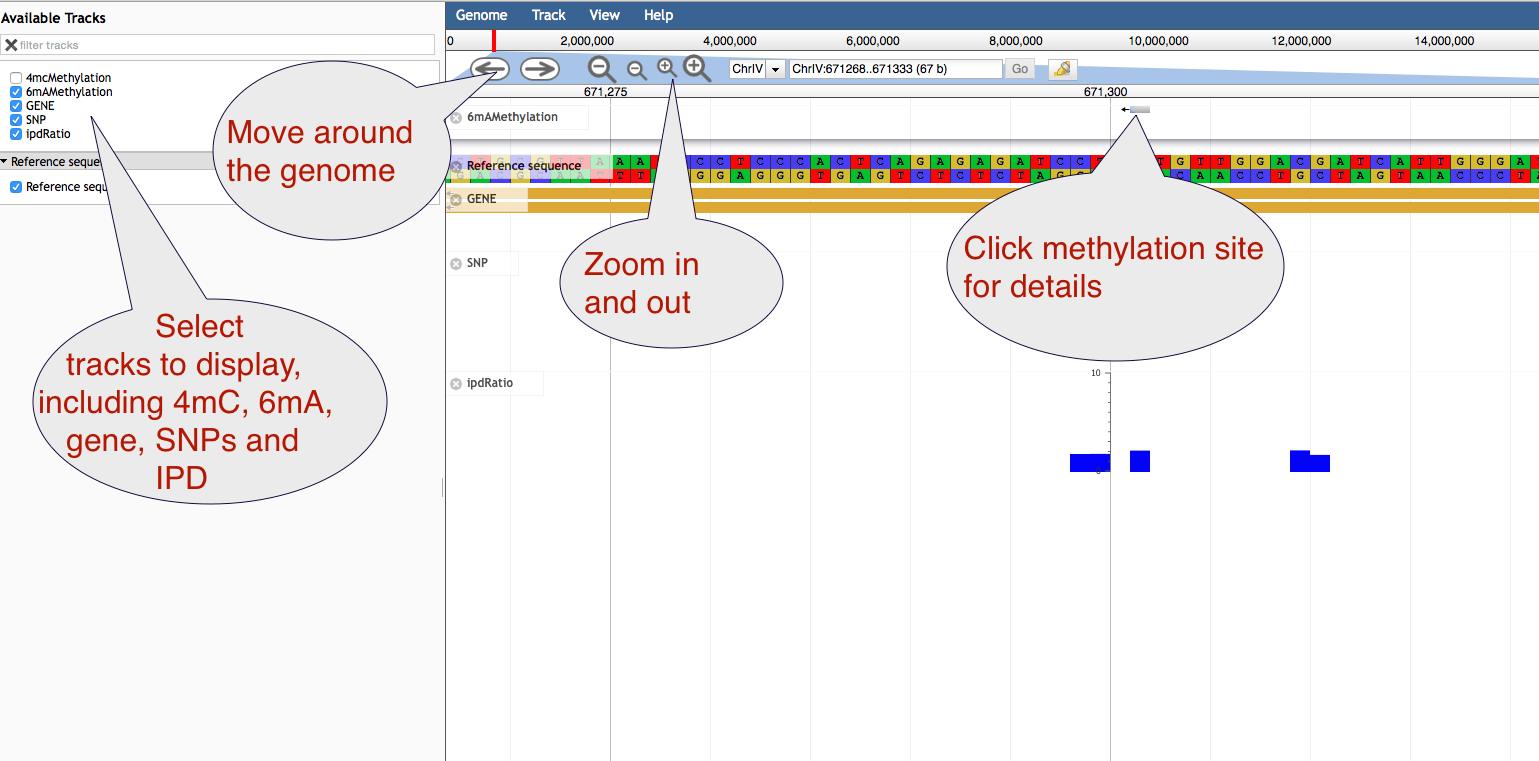
Genome Browser
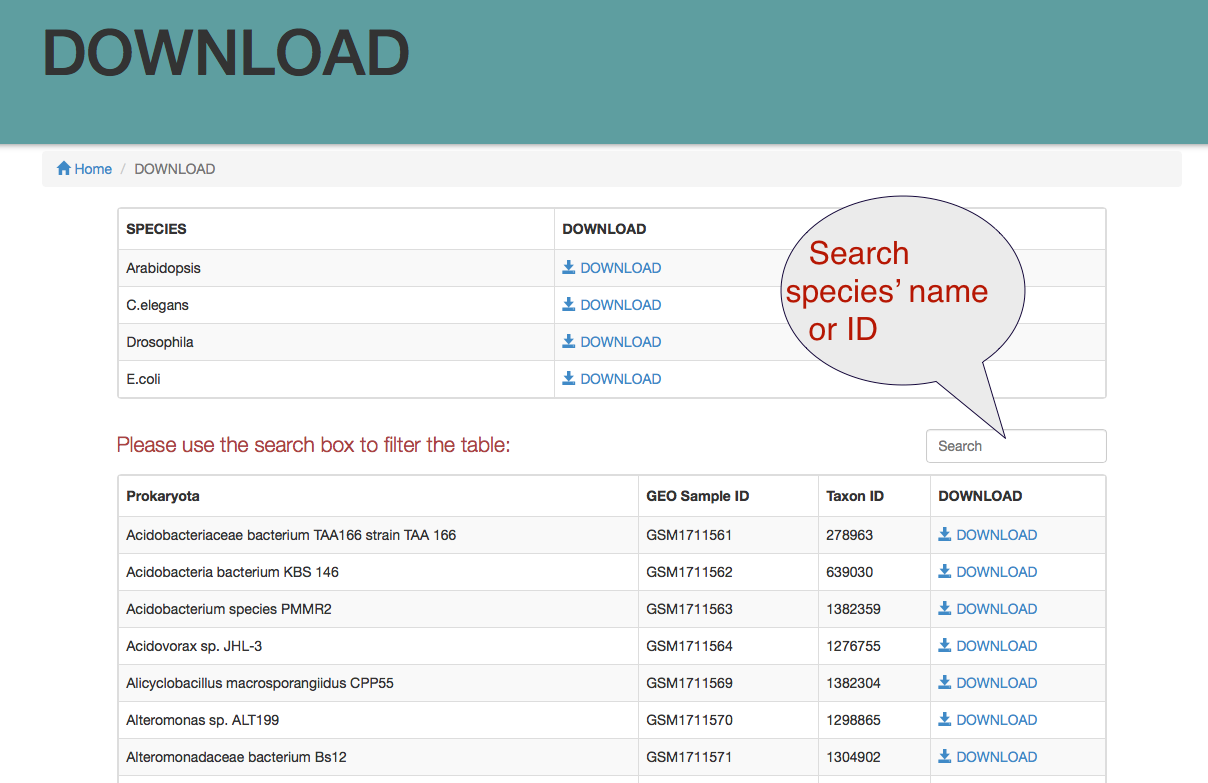
Download
News and Updates
May, 19, 2016
MethSMRT now include 6mA DNA methylation sites for 136 prokaryotes and 4 eukaryotes, including Arabidopsis, C. elegans, Drosophila and yeast.
June, 10, 2016
MethSMRT now includes 4mC DNA methylation information for 136 prokaryotes and 4 eukaryotes.
July, 28, 2016
We added 13 prokaryotes and three eukaryotes, including mouse, Diospyros lotus and Tolypocladium.
About Us
If you would like to cite our database, please use the url blow.
http://sysbio.sysu.edu.cn/methsmrt
If you have any comments or suggestions, please contact
Zhi Xie at xiezhi at gmail dot com
For technical support, please contact
PohaoYe at du.shen at hotmail dot com
Our address:
No. 54, XianlieNan Road,
Yuexiu District,
Guangzhou,
China
510060